ProductKaneka Easy DNA Extraction Kit version 2(Research reagent)
Sample categories: Microorganisms
Types of samples in detail (Microorganisms)
<Customers’ answers to our survey have confirmed DNA can be extracted from the samples below>
- Microorganisms cultured from air samples
- Mycobacterium smegmatis
- M. leprae and other antimicrobials
- BCG
- Microalgae (Raphidophyta)
- Mutans
- Bacteria (Photobacterium damselae)
- E. coli
<Samples reported in papers>
- Euglena gracilis
- M. abscessus
- A. pacificum
- A. catenella
- Lactic acid bacteria
- Streptococcus canis
- Midges/cells
- Pseudomonas aeruginosa
Survey result: extraction results and comments
This page provides answers to our survey by customers who have used Kaneka Easy DNA Extraction Kit version 2. They are published as they are.
This is not a guarantee of the accuracy or reliability of extraction results of the samples. Whether or not DNA can be extracted depends on the quantity, type and condition of each sample. If you wish to know whether this kit is suitable for your purpose, we recommend trying a free sample.
E. coli
Result: DNA was extracted.
Comment: We varied the number of bacterial cells between 1 x 107 and 5 x 108 and we were able to detect DNA from 1/5 of 5 x 107 using agarose gel stained by Midori Green Xtra. We were able to extract DNA without damage.

Acid-fast bacilli including mycobacterium smegmatis and M. leprae
Result: DNA was extracted.
Comment: As the extraction steps were very simple, we were able to minimize the variation in DNA recovery rate between samples. Considering the simplicity of the procedure, the DNA recovery rates are surprisingly high.

BCG
Result: DNA was extracted.
Comment: It was simple and easy without the need to use columns.

Usage examples
DNA extraction and amplification from Candida colonies
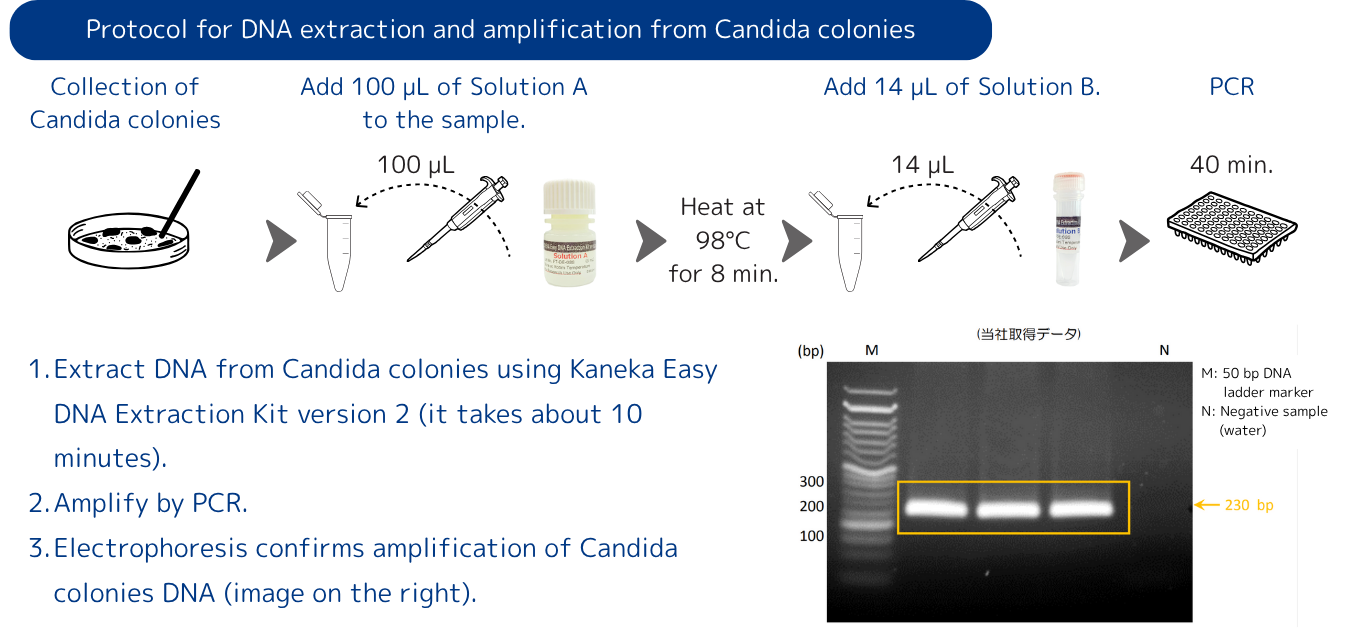
- Extract DNA from Candida colonies using Kaneka Easy DNA Extraction Kit version 2 (it takes about 10 minutes).
- Amplify by PCR.
- Electrophoresis confirms amplification of Candida colonies DNA (image on the right).
Papers using this product
- 【Mycobacteria】
Evaluation of PyroMark Q24 pyrosequencing as a method for the identification of mycobacteria - 【Euglena gracilis】
Highly Efficient CRISPR-Associated Protein 9 Ribonucleoprotein-Based Genome Editing in Euglena gracilis - 【Enterococcus faecalis】
Establishment of a polymerase chain reaction-based method for strain-level management of Enterococcus faecalis EF-2001 using species-specific sequences identified by whole genome sequences.